Highlights
Mechanistic origin of cell-size control and homeostasis in bacteria
Fangwei Si, Guillaume Le Treut, John T Sauls, Stephen Vadia, Petra Anne Levin and Suckjoon Jun
Current Biology, 29, 1-11 (2019)
[online][PDF][Google Scholar][UCSD news]
Fangwei Si, Guillaume Le Treut, John T Sauls, Stephen Vadia, Petra Anne Levin and Suckjoon Jun
Current Biology, 29, 1-11 (2019)
[online][PDF][Google Scholar][UCSD news]
Video abstract explaining the mechanistic origin of adder.
Fundamental Principles in Bacterial Physiology - History, Recent progress, and the Future with Focus on Cell Size Control: A Review
Suckjoon Jun, Fangwei Si, Rami Pugatch and Matthew Scott
Reports on Progress in Physics (2018)
[online][PDF][arXiv Preprint (PDF)]
Suckjoon Jun, Fangwei Si, Rami Pugatch and Matthew Scott
Reports on Progress in Physics (2018)
[online][PDF][arXiv Preprint (PDF)]
A comprehensive overview of bacterial physiology focusing on the fundamental problems of the field, with extensive reference to milestones that shaped the field from the inception of the field to the most up-to-date developmnent.
Invariance of initiation mass and predictability of cell size in Escherichia coli
Fangwei Si*, Dongyang Li*, Sarah E. Cox, John T. Sauls, Omid Azizi, Cindy Sou, Amy B. Schwartz, Michael J. Erickstad, Yonggun Jun, Xintian Li and Suckjoon Jun
Current Biology, 2017
[OPEN ACCESS (PDF)][Download full dataset][UCSD News][phys.org]
Fangwei Si*, Dongyang Li*, Sarah E. Cox, John T. Sauls, Omid Azizi, Cindy Sou, Amy B. Schwartz, Michael J. Erickstad, Yonggun Jun, Xintian Li and Suckjoon Jun
Current Biology, 2017
[OPEN ACCESS (PDF)][Download full dataset][UCSD News][phys.org]
We proposed the "unit cell" as the fundamental building block of cell size and its invariance underlies the predictability of cell size using the general growth law.
Cell-size control and homeostasis in bacteria
S. Taheri-Araghi, S. Bradde, J. T. Sauls, N. S. Hill, P. A. Levin, J. Paulsson, M. Vergassola, and S. Jun
Current Biology 25(3), 385–391, 2015
[online] [PDF+extended SI] [Google Scholar] [news coverage] [download data]
S. Taheri-Araghi, S. Bradde, J. T. Sauls, N. S. Hill, P. A. Levin, J. Paulsson, M. Vergassola, and S. Jun
Current Biology 25(3), 385–391, 2015
[online] [PDF+extended SI] [Google Scholar] [news coverage] [download data]
Cell-size maintenance: universal strategy revealed
S. Jun & S. Taheri-Araghi
Trends in Microbiology 23(1), 4–6, 2015
[online] [PDF] [Google Scholar]
S. Jun & S. Taheri-Araghi
Trends in Microbiology 23(1), 4–6, 2015
[online] [PDF] [Google Scholar]
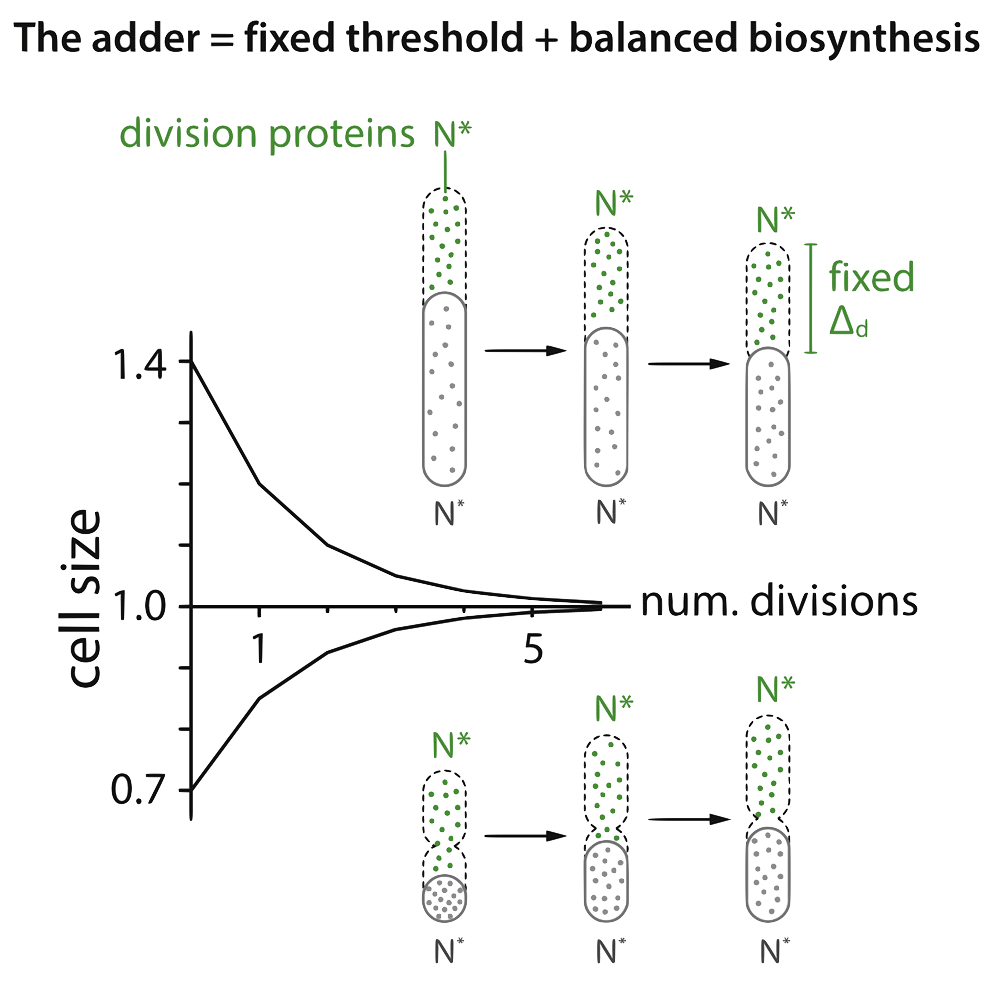
 We believe these two articles together establish a fundamental quantitative 'adder' principle for size control and homeostatis, one of the long-standing questions in biology.
We believe these two articles together establish a fundamental quantitative 'adder' principle for size control and homeostatis, one of the long-standing questions in biology.
Size convergence by the Adder principle
The multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymer
Brenda Youngren, Henrik Jork Nielsen, Suckjoon Jun, and Stuart Austin.
Genes & Development 28:71-84, 2014
[open access full article] [Google Scholar]
[see also 'The bacterial chromosome: a physical biologist’s apology. A perspective.']
Brenda Youngren, Henrik Jork Nielsen, Suckjoon Jun, and Stuart Austin.
Genes & Development 28:71-84, 2014
[open access full article] [Google Scholar]
[see also 'The bacterial chromosome: a physical biologist’s apology. A perspective.']
At long last, and for the first time, we understand the organization, dynamics, segregation of the E. coli chromosome under all growth and cell-cycle conditions in vivo.

Physical manipulation of the bacterial chromosome reveals its soft nature
James Pelletier, Ken Halvorsen, Bae-Yeun Ha, Raffaella Paparcone, Steven Sandler, Conrad Woldringh, Wesley Wong, and Suckjoon Jun
PNAS Plus 109(40), E2649-E2656, 2012.
[open access full article] [PNAS highlight] [Google Scholar]
[Nature Methods highlight]
James Pelletier, Ken Halvorsen, Bae-Yeun Ha, Raffaella Paparcone, Steven Sandler, Conrad Woldringh, Wesley Wong, and Suckjoon Jun
PNAS Plus 109(40), E2649-E2656, 2012.
[open access full article] [PNAS highlight] [Google Scholar]
[Nature Methods highlight]
All chromosomes must fold to fit within cellular containers. Though strongly confined, the bacterial chromosome exhibits highly dynamic behavior during the cell cycle. Many models have been proposed, but few measurements have quantified the essential micromechanical properties of the bacterial chromosome. In this work, we experimentally demonstrated and quantified the fundamentally soft nature of the bacterial chromosome and the entropic forces that can cause fast compaction in a crowded intracellular environment. This 7-year work, experiment and theory combined, is a quantitative demonstration of the loaded entropic spring model of the bacterial chromosome.
Entropy as the driver of chromosome segregation
Suckjoon Jun and Andrew Wright
Nature Reviews Microbiology 8, 600-607 (2010).
[online] [PDF] [Small Things Considered] [Google Scholar]
Suckjoon Jun and Andrew Wright
Nature Reviews Microbiology 8, 600-607 (2010).
[online] [PDF] [Small Things Considered] [Google Scholar]
In our PNAS 2006 article, we proposed conformational entropy- driven chromosome organization and segregation in bacteria. This work quantitatively discusses a set of minimal physical conditions the bacterial chromosomes should satisfy to segregate spontaneously, and goes beyond the original PNAS 2006 article. Furthermore, this article critically examines the existing models.
Robust growth of Escherichia coli,
Ping Wang, Lydia Rober, James Pelletier, Wei Lien Dang, Francois Taddei, Andrew Wright, and Suckjoon Jun
Current Biology 20, 1099-1103, 2010.
[online] [PDF] [F1000] [Small Things Considered]
[Google Scholar]
[The Scientist Top 7 Biology]
[The Scientist Top 7 Biochemistry]
[download data]
Ping Wang, Lydia Rober, James Pelletier, Wei Lien Dang, Francois Taddei, Andrew Wright, and Suckjoon Jun
Current Biology 20, 1099-1103, 2010.
[online] [PDF] [F1000] [Small Things Considered]
[Google Scholar]
[The Scientist Top 7 Biology]
[The Scientist Top 7 Biochemistry]
[download data]
In this work, we introduce the mother machine for the first time. The mother machine allowed us to follow thousands of individual mother cells for hundreds of consecutive generations. The results were surprising. See the movie below.
Just Published and Upcoming
|
Tools and methods for high-throughput single-cell imaging with the mother machine Ryan Thiermann, Michael Sandler, Gursharan Ahir, John T. Sauls, Jeremy W. Schroeder, Steven D. Brown, Guillaume Le Treut, Fangwei Si, Dongyang Li, Jue D. Wang, Suckjoon Jun, Elife (2024) [online] |
|
|
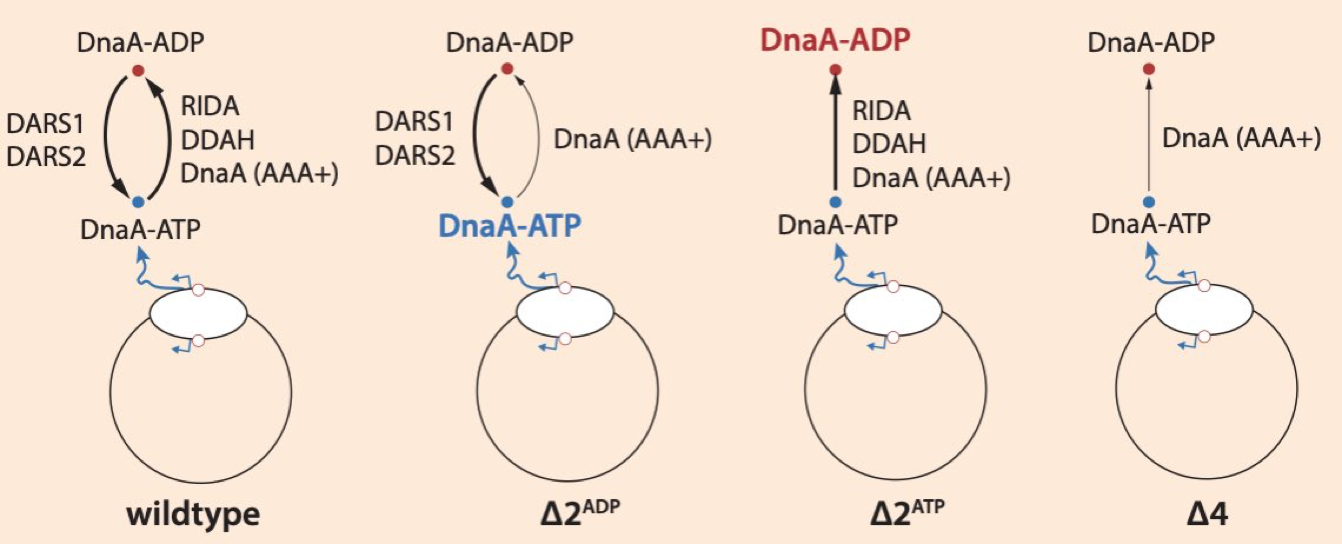
Robust control of replication initiation in the absence of DnaA-ATP ⇋ DnaA-ADP regulatory elements in Escherichia coli Thias Boesen, Godefroid Charbon, Haochen Fu, Cara Jensen, Michael Sandler, Suckjoon Jun, and Anders Lobner-Olesen, bioRxiv (2023) [online] |
|
Temperature Compensation through Kinetic Regulation in Biochemical Oscillators Haochen Fu, Chenyi Fei, Qi Ouyang, Yuhai Tu, arXiv (2024) [online] |
|
|
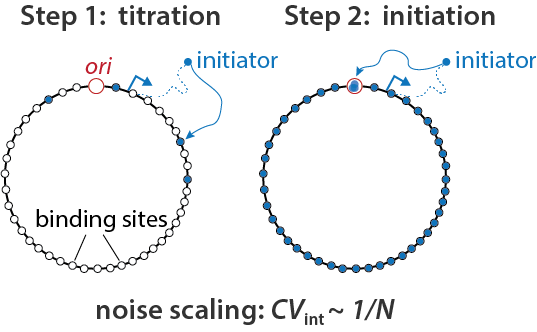
Bacterial Replication Initiation as Precision Control by Protein Counting Haochen Fu, Fangzhou Xiao, Suckjoon Jun, PRX Life (2023) [online][Viewpoint] |
|
Robust and resource-optimal dynamic pattern formation of Min proteins in vivo Ziyuan Ren, Henrik Weyer, Laeschkir Würthner, Dongyang Li, Cindy Sou, Daniel Villarreal, Erwin Frey, Suckjoon Jun, bioRxiv (2023) [online] |
|
|
Microbiology: How to Bridge Mechanisms and Phenomenology Suckjoon Jun, Editorial for a special issue of JMB (2020) [online] |
|
|
Single-cell data and correlation analysis support the independent double adder model in both Escherichia coli and Bacillus subtilis Guillaume Le Treut, Fangwei Si, Dongyang Li and Suckjoon Jun, bioRxiv (2020) [online][Jupyter notebook] |
|
|
Comment on ‘Initiation of chromosome replication controls both division and replication cycles in E. coli through a double-adder mechanism’ Fangwei Si, Guillaume Le Treut, Dongyang Li and Suckjoon Jun, bioRxiv (2020) [online][Jupyter notebook] |
|
|
Mother machine image analysis with MM3 John T. Sauls, Jeremy W. Schroeder, Steven D. Brown, Guillaume Le Treut, Fangwei Si, Dongyang Li, Jue D. Wang, Suckjoon Jun bioRxiv (2019) [online] |
|
|
Control of Bacillus subtilis Replication Initiation during Physiological Transitions and Perturbations John T. Sauls, Sarah E. Cox, Quynh Do, Victoria Castillo, Zulfar Ghulam-Jelani, Suckjoon Jun mBio, 10, e02205-19 (2019) [online][PDF] |
|
|
Mechanistic origin of cell-size control and homeostasis in bacteria Fangwei Si, Guillaume Le Treut, John T Sauls, Stephen Vadia, Petra Anne Levin and Suckjoon Jun Current Biology, 29, 1-11 (2019) [online][PDF][Google Scholar][UCSD news] |
|
Cell boundary confinement sets the size and position of the E. coli chromosome Fabai Wu, Pinaki Swain, Louis Kuijpers, Xuan Zheng, Kevin Felter, Margot Guurink, Jacopo Solari, Suckjoon Jun, Thomas S. Shimizu, Debasish Chaudhuri, Bela Mulder* and Cees Dekker* Current Biology, 29, 2131-2144.e4 (2019) [online] |
|
|
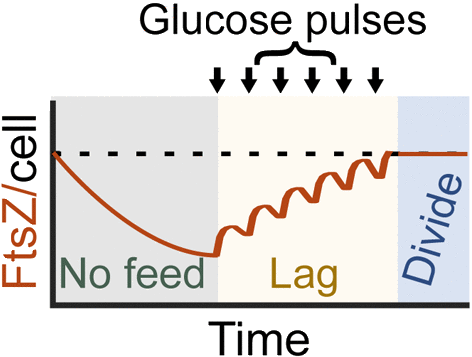
Synthesis and degradation of FtsZ quantitatively predicts the first cell division in starved bacteria K. Sekar, R. Ruscon, JT Sauls, T. Fuhrer, E. Noor, J. Nguyen, VI Fernandez, MF Buffing, M. Berney, S. Jun, R. Stocker, U. Sauer Mol. Sys. Biol., (2018) 14, e8623 [online][PDF][Google Scholar][ETHZ news] |
|
Promoting an "Auteur Theory" for Young Scientists: Preserving Excitement and Creativity Arshad Desai and Suckjoon Jun BioEssays, 1800147, (2018) [PDF] |
|
|
Fundamental Principles in Bacterial Physiology - History, Recent progress, and the Future with Focus on Cell Size Control: A Review Suckjoon Jun, Fangwei Si, Rami Pugatch and Matthew Scott Reports on Progress in Physics (2018) [online][PDF][Google Scholar][arXiv Preprint (PDF)] |
|
|
Protocol for Construction of a Tunable CRISPR Interference (tCRISPRi) Strain for Escherichia coli Xintian Li, Cindy Sou, and Suckjoon Jun BIO-PROTOCOL 7(19):e2574 (2017). [online] |
|
|
A fundamental unit of cell size in bacteria Suckjoon Jun and Michael Rust TRENDS in Genetics 33, 433-435 (2017) [online][PDF] |
|

|
Invariance of initiation mass and predictability of cell size in Escherichia coli Fangwei Si*, Dongyang Li*, Sarah E. Cox, John T. Sauls, Omid Azizi, Cindy Sou, Amy B. Schwartz, Michael J. Erickstad, Yonggun Jun, Xintian Li and Suckjoon Jun Current Biology (2017) [OPEN ACCESS (PDF)][Download full dataset][Google Scholar][UCSD News][phys.org][ScienceDaily][KENNISLINK (in Dutch!)][Commentary in Current Biology] |
|
tCRISPRi: tunable and reversible, one-step control of gene expression Xintian Li, Yonggun Jun, Michael J. Erickstad, Steven D. Brown, Adam Parks, Donald L. Court, and Suckjoon Jun Scientific Reports 6, 39076 (2016) [PDF][Google Scholar][strain request] |
|
|
Adder and a coarse-grained approach to cell size homeostasis in bacteria John T Sauls, Dongyang Li, and Suckjoon Jun Current Opinion in Cell Biology 38:38-44 (2016) [online][PDF][Google Scholar] |
|
|
Self-Consistent Examination of Donachie's Constant Initiation Size at the Single-Cell Level Sattar Taheri Frontiers in Microbiology 6:1349 (2015) [online] |
|
|
Single-cell cultivation in microfluidic devices Sattar Taheri-Araghi and Suckjoon Jun Hydrocarbon and Lipid Microbiology Protocols, Springer Protocols Handbooks, doi:10.1007/8623_2015_68 (2015) [online] |
|
|
Single-cell physiology Sattar Taheri, Steven Brown, John T. Sauls, Dustin McIntosh, Suckjoon Jun Annual Review of Biophysics 44, 123-142 (2015) [download] [Google Scholar] |
|
|
Complete Genome Sequence of Escherichia coli NCM3722 S. D. Brown and S. Jun Genome Announc 3(4):e00879-15. doi:10.1128/genomeA.00879-15. [online][Google Scholar][strain request] |
|

|
Cell-size control and homeostasis in bacteria S. Taheri-Araghi, S. Bradde, J. T. Sauls, N. S. Hill, P. A. Levin, J. Paulsson, M. Vergassola, and S. Jun Current Biology 25(3), 385–391, 2015 [online] [PDF+extended SI] [Google Scholar] [news coverage] [download data] |
|
Cell-size maintenance: universal strategy revealed S. Jun & S. Taheri-Araghi Trends in Microbiology 23(1), 4–6, 2015 [online] [PDF] [Google Scholar] |
|
|
Chromosome, cell cycle, and entropy S. Jun. Biophysical Journal 108(4), 785-786, 2015 [online] [PDF] [from the cover] |
Publications
[La Jolla, CA, 2013-] [Cambridge, MA, 2007-2012] [Paris & Amsterdam, 2004-2007] [Vancouver, BC, 1999-2004]
| [La Jolla, CA, 2013-] | |
|---|---|
|
Q: How does E. coli control its shape?Bending forces plastically deform growing bacterial cell wallsAriel Amir, Farinaz Babaeipour, Dustin McIntosh, David Nelson, and Suckjoon Jun Proc. Nat. Acad. Sci. USA 111, 5778-5783, 2014 [open access full article] [News in Nature Physics] [Google Scholar] |

|
See highlights on the rightThe multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymerBrenda Youngren, Henrik Jork Nielsen, Suckjoon Jun, and Stuart Austin. Genes & Development 28:71-84, 2014 [open access full article] [Google Scholar] |
| [Cambridge, MA, 2007-2012] | |

|
See highlights on the rightPhysical manipulation of the Escherichia coli chromosome reveals its soft natureJames Pelletier, Ken Halvorsen, Bae-Yeun Ha, Raffaella Paparcone, Steven Sandler, Conrad Woldringh, Wesley Wong, and Suckjoon Jun PNAS Plus 109(40), E2649-E2656, 2012. [open access full article] [PNAS highlight] [Nature Methods highlight] [Google Scholar] |

|
Q: "superblobs"?Intrachain ordering and segregation of polymers under confinementY. Jung, J. Kim, S. Jun and B.-Y. Ha Macromolecules 45 (7), 3256-3262, 2012. [online] [PDF] [Google Scholar] |

|
Q: What is the behavior of two ring polymers in strong confinement?Ring polymers as model bacterial chromosomes: confinement, chain topology, single chain statistics and how they interact.Y. Jung, C. Jeon, J. Kim, H. Jeong, S. Jun and B.-Y. Ha Soft Matter 8, 2095-2102, 2012. [online] [PDF] [Google Scholar] |

|
See highlights on the rightEntropy as the driver of chromosome segregation.Suckjoon Jun and Andrew Wright Nature Reviews Microbiology 8, 600-607 (2010). [online] [PDF] [Small Things Considered] [Google Scholar] |

|
See highlights on the rightRobust growth of Escherichia coliPing Wang, Lydia Rober, James Pelletier, Wei Lien Dang, Francois Taddei, Andrew Wright, and Suckjoon Jun Current Biology 20, 1099-1103, 2010. [online] [PDF] [F1000] [Google Scholar] [Small Things Considered] [The Scientist Top 7 Biology] [The Scientist Top 7 Biochemistry] [download data] |
|
Q: What is the typcal timescale of relaxation of a chain in a cylinder?A self-avoiding polymer trapped inside a cylindrical pore: Flory free energy and unexpected dynamicsYoungkyun Jung, Suckjoon Jun, Bae-Yeun Ha Phys. Rev. E 79, 061912 (2009). [PDF] [Google Scholar] |

|
Viewpoint article for Physics (APS)Just-in-time DNA replicationSuckjoon Jun and Nick Rhind Physics 1, 32 (2008). [online] [Google Scholar] |
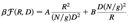
|
Q: How to fix Flory’s mistake to obtain forcecompression curves in a cylindrical pore?Compression and stretching of a self-avoiding chain in cylindrical nanoporesSuckjoon Jun, D. Thirumalai and Bae-Yeun Ha Phys. Rev. Lett. 101, 138101 (2008). [PDF] [Google Scholar] |
| [Paris & Amsterdam, 2004-2007] | |
|
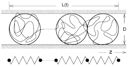
Q: What is the real relaxation time of a selfavoiding chain in a cylinder?Unexpected relaxation dynamics of a self-avoiding polymer in cylindrical confinementAxel Arnold, Behnaz Borzorgui, Daan Frenkel, Bae-Yeun Ha and Suckjoon Jun J. Chem. Phys. 127, 164903 (2007). [online] [PDF] [Google Scholar] |

|
Q: Is entropy-driven segregation in a cylinder fast enough?Timescale of entropic segregation of flexible polymers in confinement: Implications for chromosome segregation in filamentous bacteriaAxel Arnold and Suckjoon Jun Phys. Rev. E 76, 031901 (2007). [online] [PDF] |
|
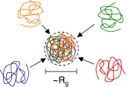
Q: What is the free energy of arbitrary number of overlapping self-avoiding chains?Confined space and effective interactions of multiple self-avoiding chainsSuckjoon Jun, Axel Arnold and Bae-Yeun Ha Phys. Rev. Lett. 98, 128303 (2007) [online] [PDF] [Google Scholar] |

|
This papers introduces two novel ideas on bacterial chromosomes. 1. Conformational entropy can drive strongly confined DNA demix during replication, 2. Newly-replicated DNA will be extruded to the periphery of the bacterial chromosome in vivo.Entropy-driven sptial organization of highly confined polymers: Lessons for the bacterial chromosome.Suckjoon Jun and Bela Mulder PNAS 103, 12388 (2006) [online] [F1000] [JCB highlight] [Google Scholar] |
| [Vancouver, BC, 1999-2004] | |
|
Ch. 4. Semiflexible polymers: from statics to dynamics Jun, S., Bechhoefer J., and Ha, B.-Y. Edited by Pu Chen (Woodhead Publishing Ltd, Cambridge, UK) (July, 2005) [online] |
|
|
Kinetic model of DNA replication and the looping of semiflexible polymers Suckjoon Jun PhD Thesis (2004) [PDF] |

|
Nucleation and growth in one dimension part II: Application to DNA replication kinetics Suckjoon Jun and John Bechhoefer (Phys.Rev.E 71, 011909 (2005); cont-mat/0408297) [PDF] [Google Scholar] |
|
Nucleation and growth in one dimension part I: The generalized Kolmogorov-Johnson-Mehl-Avrami model Suckjoon Jun, Haiyang Zhang, and John Bechhoefer (Phys.Rev. E 71, 011908 (2005); cont-mat/0408260) [PDF] [Google Scholar] |

|
Self-Assembly of the Ionic Peptide EAK16: the effect of charge distributions on self-assembly S. Jun, Y. Hong, H. Imamura, B.-Y. Ha, J. Bechhoefer, and P. Chen Biophys. J. 87, 1249-1259 (2004) [PDF] [Google Scholar] |

|
Persistence length of chromatin determines origin spacing in Xenopus early-embryo DNA replication: Quantitative comparisons between theory and experiment Suckjoon Jun, John Herrick, Aaron Bensimon, and John Bechhoefer Cell Cycle 3(2), 223-229 (2004) [PDF] [Google Scholar] |
|
Diffusion-limited loop formation of semiflexible polymers: Kramers theory and the intertwined time scales of chain relaxation and closing Suckjoon Jun, John Bechhoefer, and Bae-Yeun Ha Europhys. Lett. 64(3), 420-426 (2003) [PDF] [Google Scholar] |
|
Role of Polymer Loops in DNA replication Suckjoon Jun and John Bechhoefer Physics in Canada 59(2), pp. 85-92 (2003) [PDF] [Google Scholar] |
|
Kinetic model of DNA replication in eucaryotic organisms John Herrick, Suckjoon Jun, John Bechhoefer, and Aaron Bensimon J. Mol. Biol. 320, 741-750 (2002) [PDF] [Google Scholar] |
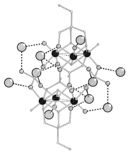
|
Heisenberg spin-triangle in {V6}-type magnetic molecules: Experiment and theory M Luban, F Borsa, S Budko, P Canfield, S Jun, JK Jung, P Kögler, D Mentrup, A Müller, R Modler, D Procissi, BJ Suh, and M Torikachvili Phys. Rev. B 66, 054407 (2002) [PDF] [Google Scholar] |